4. Output
The main output file is called vectri.nc which is found in the run directory (unless you chose to rename it with the -o option).
The file is in standard netcdf format and you can use two commands to examine its content ncview and ``ncdump <<https://www.unidata.ucar.edu/software/netcdf/workshops/2011/utilities/Ncdump.html>``_.
4.1. NCDUMP
This command lets us know the dimensions of the output, gives a list of the model output variables, as well as their metadata. We will be using it a lot to interrogate netcdf files!
Tip
ncdump outputs the entire contents of a file, perhaps a little more than you bargained for! So use the useful option is -h to examined the file header.
Remember, if you don’t know what a netcdf file is, check out our online video introduction.
So let’s try examining the output using this command
ncdump -h vectri.nc
Tip
sometimes the header is very long - to save yourself having to scroll up and down, it is helpful to pipe the output of ncdump into the command less .. code-block:
ncdump -h vectri.nc | less
The output shows the dimensions first, and then the dump provides a list of all the model run global attributes. Here you find stored all the parameter settings used to run the model. We will see how to change this in the Command Line Options section, while the parameters themselves are outlined in the VECTRI Parameters section. Finally the output fields follows, which are grouped under the titles of vector, disease, interventions, hydrology, and input, which will be introduced next.
Note
The exact github tag release used is stored in the global attributes, along with the run command and all the simulation parameter settings. This is to ensure that, even if the model is upgraded, you should be able to download the exact code version and reproduce your results later, essential for reeproducability of published results!
4.2. NCVIEW
We can now open the output using the ncview utility. Try this out now!
ncview vectri.nc
This should open a window with the available output groups, looking like this:
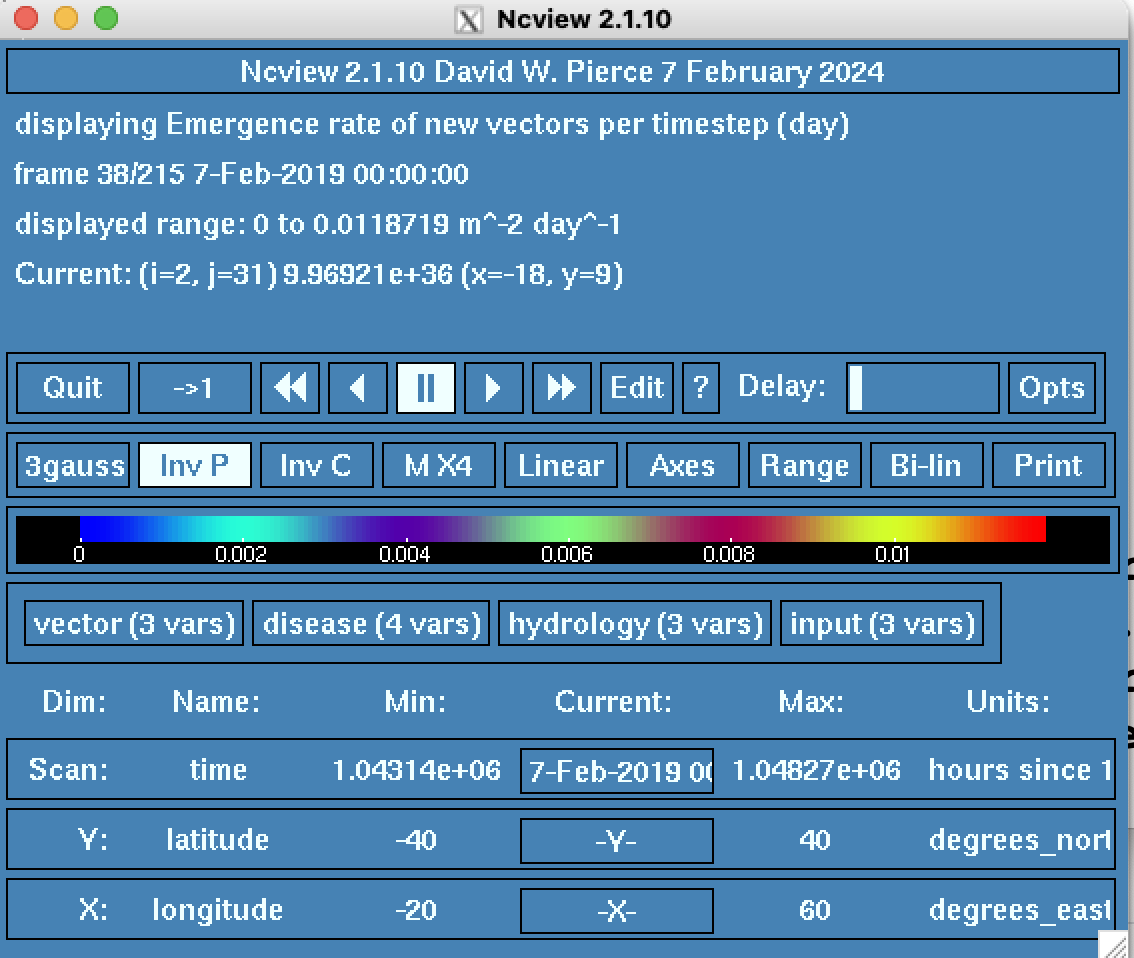
You can see that the output is divided into key groups, which in the default run consist of vector, disease, hydrology and input. If you are using interventions, there will also be a 5th group call interventions.
4.3. Output Variables
We introduce the VECTRI output for each group in turn as well as the switch to control if the output is produced. We will show you how to change that setting in the Command Line Options section.
Name |
Description |
Units |
Switch name |
Default |
|---|---|---|---|---|
vector |
vector density |
m -2 |
loutput_vector |
.true. |
larvae |
larvae density |
m -2 |
loutput_larvae |
.false. |
lbiomass |
larvae biomass |
mg m -2 |
loutput_lbiomass |
.false. |
hbr |
human bite rate |
day -1 person -1 |
loutput_hbr |
.false. |
eggs |
Density of new eggs laid per day |
m -2 day -1 |
loutput_egg |
.false. |
emergence |
Emergence rate of new vectors |
m -2 day -1 |
loutput_emergence |
.false. |
vecthostratio |
vector to host ratio |
loutput_vecthostratio |
.false. |
Name |
Description |
Units |
Switch name |
Default |
|---|---|---|---|---|
PR |
Parasite Ratio (malaria) |
loutput_pr |
.false. |
|
PRd |
Detectable Parasite Ratio (malaria) [assumes ten days before positive test] |
loutput_prd |
.true. [IVEC=0:9] |
|
cspr |
circumsporozoite protein rate - proportion of infective vectors (malaria) |
loutput_cspr |
.false. |
|
eir |
Entomological Inoculation Rate (malaria) |
day -1 person -1 |
loutput_cspr |
.true. [IVEC=0:9] |
cases |
Symptomatic cases (malaria) |
day -1 person -1 |
loutput_cases |
.false. [IVEC=0:9] |
immunity |
Proportion of population with immunity (malaria) |
loutput_immunity |
.false. |
Name |
Description |
Units |
Switch name |
Default |
|---|---|---|---|---|
sit_males |
SIT vector density |
m -2 |
loutput_sit |
.false. (switched on if intervention found) |
sit_females |
vector density of females that had bred with a SIT male |
m -2 |
loutput_sit |
.false. |
Name |
Description |
Units |
Switch name |
Default |
|---|---|---|---|---|
wperm |
Fractional coverage of breeding sites on boundaries of permanent water bodies |
fraction |
loutput_wperm |
.false. |
wurbn |
Fractional coverage of breeding sites in urban environment, not rain fed |
fraction |
loutput_wurbn |
.false. |
wpond |
Fractional coverage of temporary rain-fed breeding sites (rural puddles/ponds and urban rainfed sites) |
fraction |
loutput_wpond |
.false. |
infiltration |
Soil Infiltration Rate |
mm day -1 |
loutput_soilinfil |
.false. |
Name |
Description |
Units |
Switch name |
Default |
|---|---|---|---|---|
population_density |
population density |
m -2 |
loutput_population |
.true. |
RAIN (a) |
Precipitation |
mm day -1 |
loutput_rain |
.true. |
TEMPERATURE (a) |
Two meter temperature |
deg C (b) |
loutput_t2m |
.true. |
Note
note that rain and temperature output names are taken from the driver file used for input and not changed
temperature units are converted automatically to deg C if the input is in Kelvins.
Tip
For some purposes you may wish to have the output file “flattened”, that is, without the group hierarchical structure. For example, CDO does not work on netcdf4 files with the group structure currently. In this case you will need to convert the output to a netcdf3 file format which is easy to do with nco: ncks -3 vectri.nc vectri_3.nc. If you open the new created file vectri_3.nc in ncview you will notice that the group structure is gone and the variables are contained in a single simple block.